Transduction and Lambda Red Overview
by Pallabi Roy Chakravarty, Ph.D.

by Pallabi Roy Chakravarty, Ph.D.
Transduction is a method of bacterial gene transfer mediated by viruses. It is a type of horizontal gene transfer because it occurs from the surrounding cells or environment rather than from the parents to offspring (vertically).
To learn more about horizontal and vertical gene transfer, a brief outline of this process is found just under the table of contents in this article.
Here, you will learn about how physiological processes and resources (enzymes) in viruses are used for genetic engineering of bacteria.
Specifically, we will first discuss the details of the transduction process as it occurs in nature. Thereafter, we will describe how this process is optimized by scientists to be used as an effective gene transfer tool in bacteria.
Historical fact: Transduction was discovered by scientists Joshua Lederberg and Norton Zinder during their work at the University of Wisconsin-Madison and was first reported in the bacterium Salmonella.
Introduction to bacteriophages
Option 1: Hijacking the host cell’s machinery leading to a lytic cycle
Option 2: Viral integration without lysis
Transduction for bacterial genetic engineering in the laboratory
Lambda Red Mediated Recombineering
Transduction is the process of gene transfer where viruses act as the vector, or gene transportation vehicle. For this process a special type of virus is required, which infects bacterial cells. These viruses are known as “bacteriophages.”
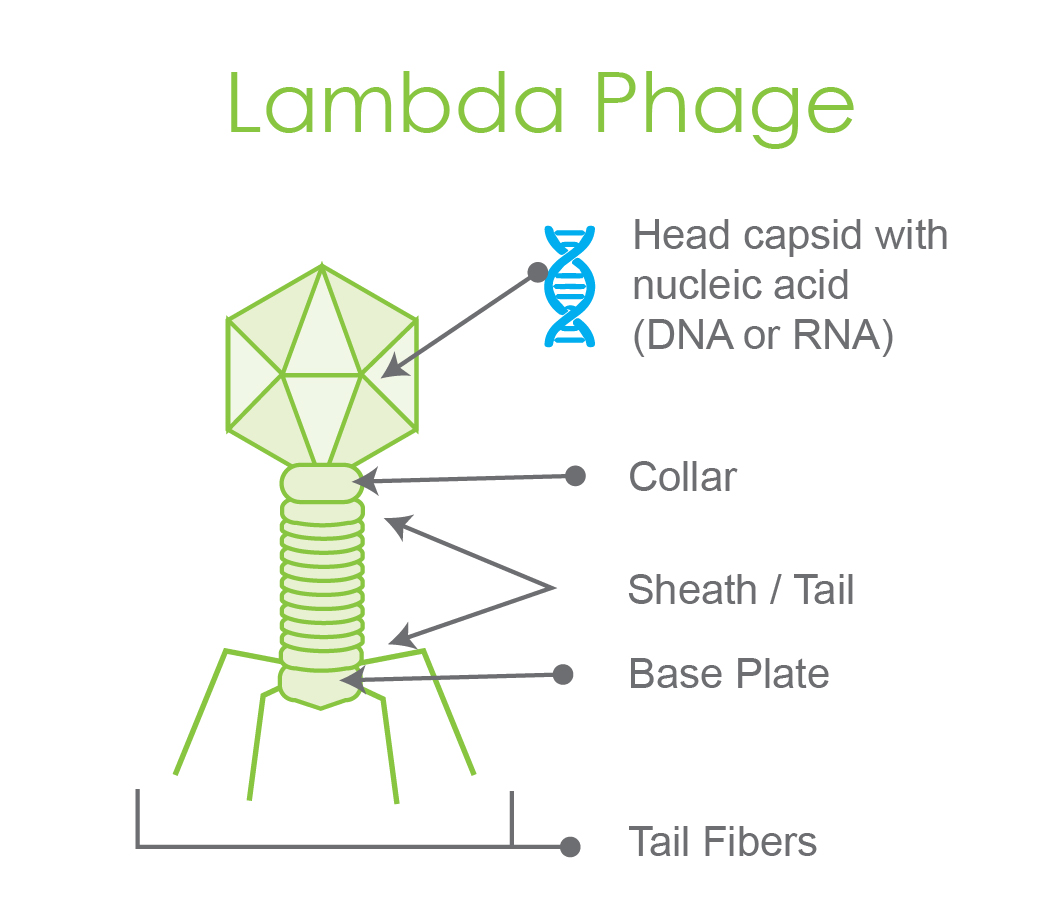
Figure 1. Pictorial representation of a labeled lambda phage, a model bacteriophage.
Bacteriophages are the mediators of transduction. As described briefly in our bacterial transformation article and depicted in figure 2 below, once a bacteriophage infects a bacterial cell, the first step is injecting its genetic material into the host cell’s cytoplasm.
The infection now can adopt one of two options:
The gene replication and expression systems of the host bacterial cell are hijacked by the phage; these physiological systems are now used for replication and expression of the injected phage genome. This leads to the synthesis of new phage particles. During the hijacking process of new phage synthesis, parts of the host DNA may also get packed in the bacteriophage capsids. As a result, this forms the first step of the gene transfer process.
Subsequently, these newly formed phages lyse the host cell and emerge out into the extracellular environment. Since lysis of the host is involved in this mechanism, it is called the “lytic cycle”. Following this, they go on to infect surrounding bacteria.

Figure 2: Lytic cycle. 1) Phage infects the host cell and injects its genome into the host cytosol. 2) Phage genes are synthesized and expressed leading to more phage particle production. The host genome is chopped off. 3) Donor cell lysis by lytic phage. The host cell is ultimately lysed with numerous phage particles bursting out of it. While most phage particles have their own genetic element (in pink), 0.1% of phages have part of the host cell’s DNA (in green). These are the transducing particles. This makes the host cell the “donor”. 4) Recipient cell infected with a transducing particle. 5) Homologous recombination between transduced donor DNA fragment and the corresponding recipient locus. 6) Transductant.
Among the newly formed emerging phages, a few may pack parts of the host genome in their capsids instead of their own genetic material. This gives rise to an interesting scenario in the second infection cycle.
Bacteriophages having their own genetic material might infect surrounding bacterial cells. This results in the same lytic cycle.
However, there will be a few bacterial cells that are infected by phages having packed part of the previous host’s DNA instead of the viral genetic element in their capsids.
Such phages are known as transducing particles. Once a transducing phage infects a second host bacterium, the previous host’s DNA is injected into its cytoplasm instead of the viral genes.
The DNA fragment from the donor (first host) now recombines with the genome of the current host (recipient).
This comprises the second and final event in the gene transfer process from one bacterium (first host that underwent lysis, this cell is the donor) to another (the second host, which is the recipient in this case).
The recipient host cell that received genetic material via transduction is called a “transductant.”
The second option occurs when injected viral genes integrate with the host cell’s genome without causing lysis of the host.
These viral genes incorporated within the host bacterial chromosomal DNA are called “prophages”(as shown in figure 3).
This type of virus-mediated gene transfer is known as the “lysogenic cycle.”
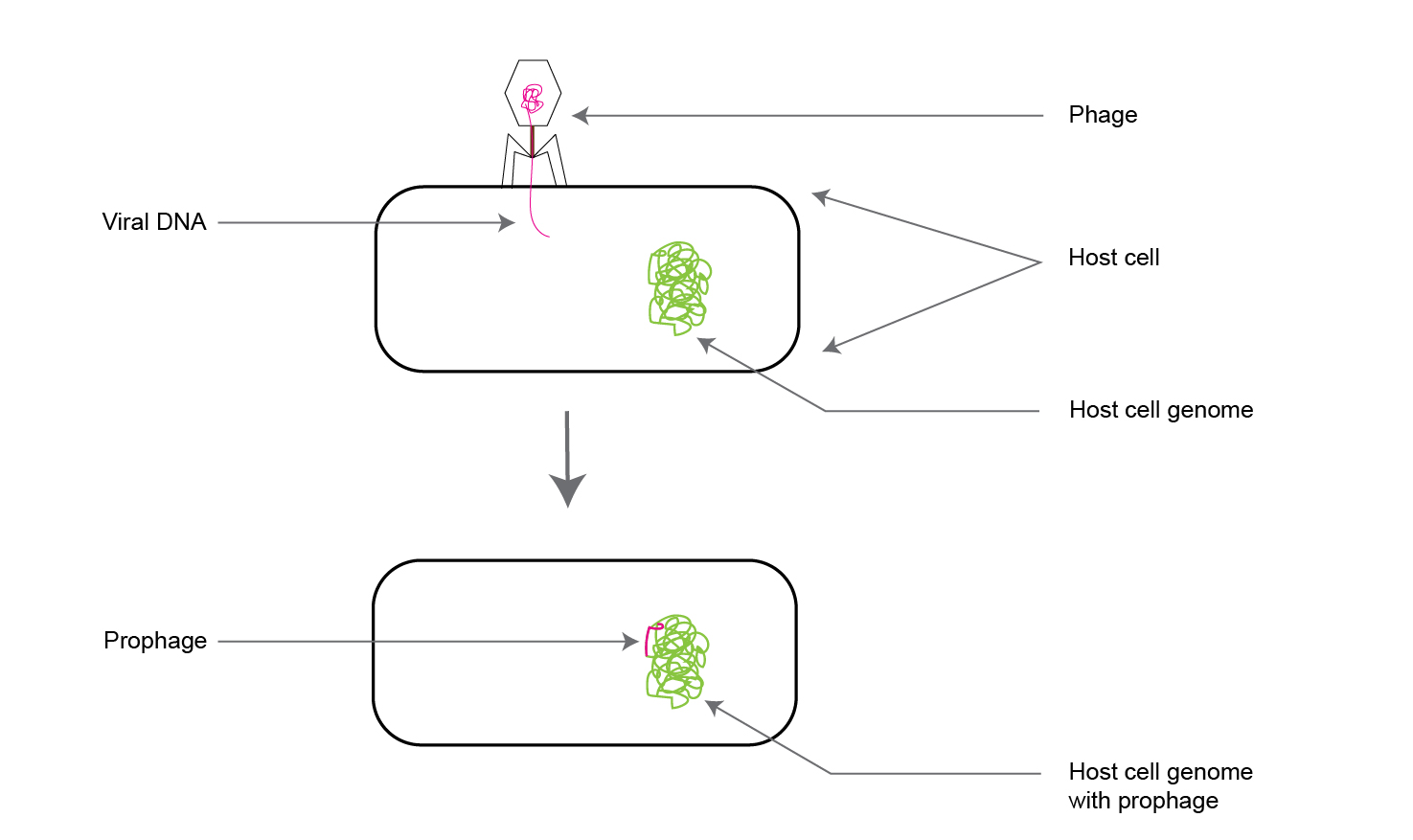
Figure 3. Illustration of a viral gene incorporating into a host cell genome, which is called a “prophage.”
When the infected bacterium divides, the prophage is also transmitted to the progeny daughter cells along with the rest of the parent cell’s genomic DNA.
Such daughter cells with the prophage are called “lysogens.”
Interestingly, both horizontal and vertical gene transmission is involved in this viral reproductive mechanism.
When the viral genes integrate into the parent cell’s DNA as a prophage, it is horizontal transfer. However, when that same prophage is transmitted from the parent to the daughter progeny, it is vertical transmission.
Integrated prophages can passively be vertically transmitted for multiple generations. But, under certain circumstances, they can switch to the lytic cycle.

Figure 4: Lysogenic cycle
Interesting Note: Bacteriophages that are capable of undergoing only the lytic cycle are called “virulent phages” while those that can switch between both lifestyles are “temperate phages.”
In both academic and industrial laboratories, transduction is a regular method to genetically modify bacteria.
Like using a plasmid, a DNA fragment of interest can be cloned in a viral vector and introduced into a suitable recipient cell. Further, using transduction, a gene / mutation from one bacterial strain can be transmitted to a bacterial strain of choice.
For this purpose, the bacteriophage P1 is one of the most used viral tools. Using lytic cycle, a fragment of interest from the donor bacterial strain’s genome is transduced into the chromosome of a suitable recipient strain.
The required transduction mode is lytic; however, the P1 phage found in nature is temperate. Therefore, when the P1 phage infects the donor strain, either lytic or lysogenic cycles might be adopted.
In P1 lysogens, the phage genes are maintained as a plasmid rather than by integrating with the host’s chromosome.
To restrict the infection cycle exclusively to the lytic form, a vir mutation has been made by scientists creating the phage variant P1vir, which is the actual bacteriophage used regularly in the lab for transduction procedures.
This P1 variant can infect only through the lytic cycle.
Scientists, over the years, have learned to exploit many aspects of bacteriophage-mediated methods of genetic engineering in addition to transduction.
One prominent example is genetic engineering in bacteria using physiological tools (enzymes) from the ‘Lambda’ bacteriophage, which facilitate a gene exchange process known as homologous recombination.
This process is known as ‘recombineering’ (recombination-mediated genetic engineering).
The recA gene in E. coli is central to the bacterium’s capability of homologous recombination.
Almost 50 years ago, scientists figured out they could use certain proteins of the bacteriophage Lambda to achieve homologous recombination, even in recA mutant E. coli strains.
This new DNA recombination system, composed of Lambda phage proteins, was named “Red” as an abbreviation of “recombination defective” to highlight uniqueness from E. coli’s own recombination machinery.
The general workflow for creating a selectable mutation in the model laboratory bacterium E. coli (and in some other bacterial strains as well) is as follows:
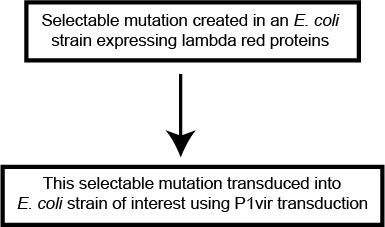
Figure 5: Schematic workflow showing how lambda red proteins and P1vir transduction are used in conjunction.

Handles are common in everyday life. We use them to help us open doors, cabinets, or to hold coffee mugs, cookware, and tools. They are...

Labeling antibodies with biotin or a fluorophore enables scientists to detect, track, and quantify that antibody and the molecules that it binds to. We cover...

Covalently conjugating a small molecule to an antibody’s surface is a process called antibody “labeling.” Labeling antibodies with small molecules such as biotin or fluorophores...

DNA gel electrophoresis is one of the most widely used analytical techniques in molecular biology, providing a simple and reliable way to separate DNA fragments...
