Plant RNA Extraction Overview: Methods, Tips, Steps and More
by Adriana Gallego, Ph.D.

by Adriana Gallego, Ph.D.
RNA plays an essential role in regulation and gene expression, and functions during plant growth and development. Therefore, obtaining sufficient and high-quality plant RNA yields is essential to many biological scientific fields.
Plant RNA has several characteristics which make its extraction challenging. In this article, we will discuss those characteristics, the types of RNA, the main steps of a plant RNA extraction, how to quantify the yield and verify the quality of the plant RNA as well as some tips for better plant RNA extraction.
Why is it so difficult to extract plant RNA?
Methods of extraction for plant RNA
Main Steps in the plant RNA extraction
Key reagents for plant RNA extraction
How to determine the quantity and quality of a plant RNA extract
7 Tips for Plant RNA Extraction
RNA is the abbreviation for the molecule Ribonucleic Acid. RNA is a very important molecule for every organism. It is composed of four types of ribonucleotide bases called adenine (A), cytosine (C), guanine (G), and uracil (U). Sometimes, RNA is compared to having a copy from a reference book (in this case from the DNA).
RNA is a single-strand molecule and contains ribose sugars (rather than deoxyribose sugars present in the DNA). Ribose sugars contain hydroxyl groups represented as OH. Those hydroxyls make RNA more reactive and susceptible to hydrolysis, causing instability of the molecule and make it more prone to degradation. Due to its instability, you must be more careful in order to manipulate RNA.
.png)
In plants, you can recognize three main types RNA:

The proportion of these three types of RNA varies in each cell: mRNA is about 2.0-2.5% of the total RNA, tRNA is about 14%, while the vast majority is occupied by rRNA (about 80%). As you will see later, these proportions play an important role in determining the quality of an RNA extraction.
In addition to the RNAs listed above, researchers have found a lot of other types of RNAs present in both plants and animals.
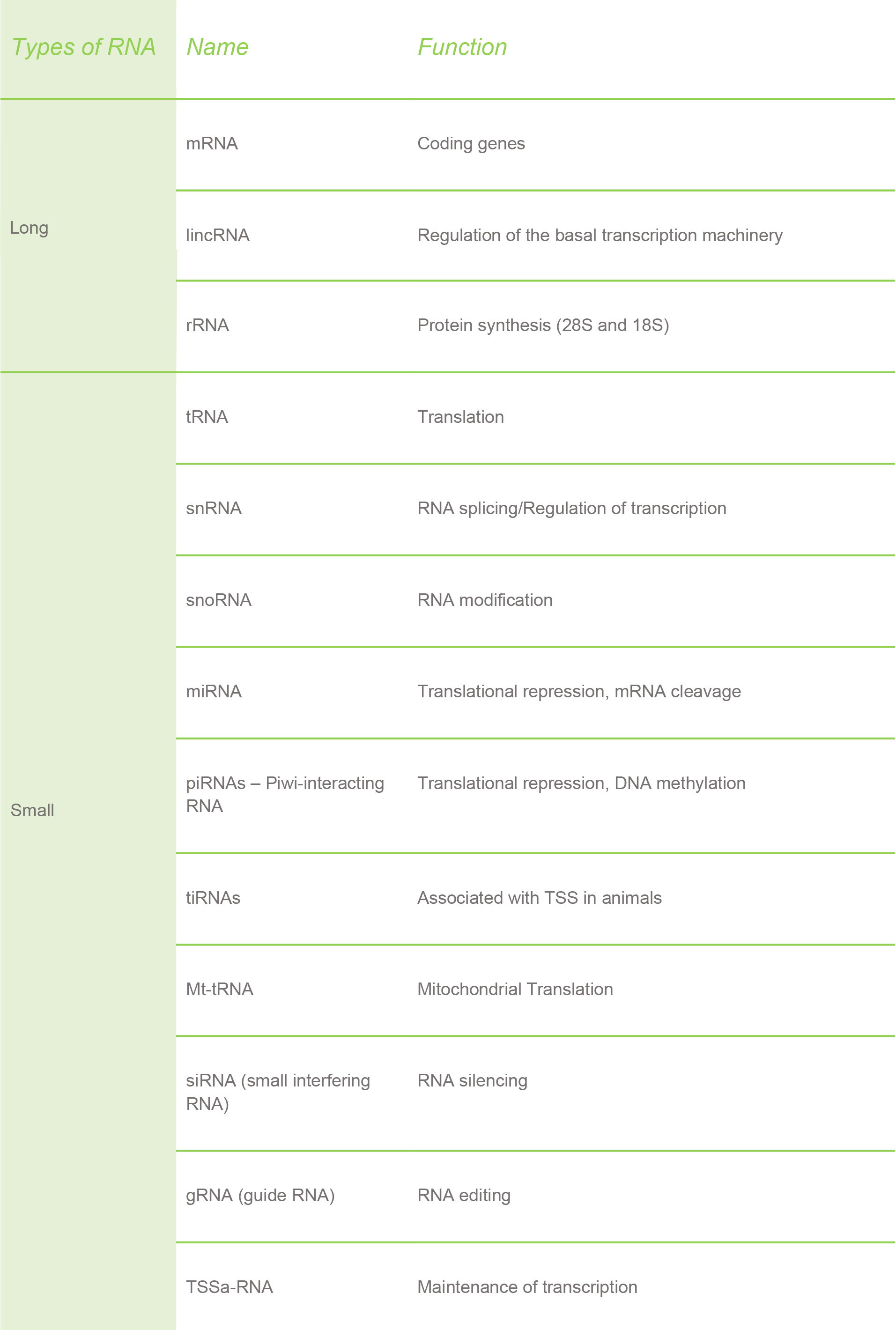
Furthermore, there are some RNAs that are specific only for plants, like the chloroplast RNA.
Similar to how RNA can be found in mitochondria, there is also RNA present in chloroplasts. The RNA in these organelles (the mitochondria and chloroplast) is explained by the endosymbiotic theory. This theory states the mitochondria and chloroplast in eukaryotic cells were once aerobic bacteria that were ingested by large anaerobic bacteria. As a consequence, plants have three completely different RNAs: nuclear, mitochondrial and chloroplast.
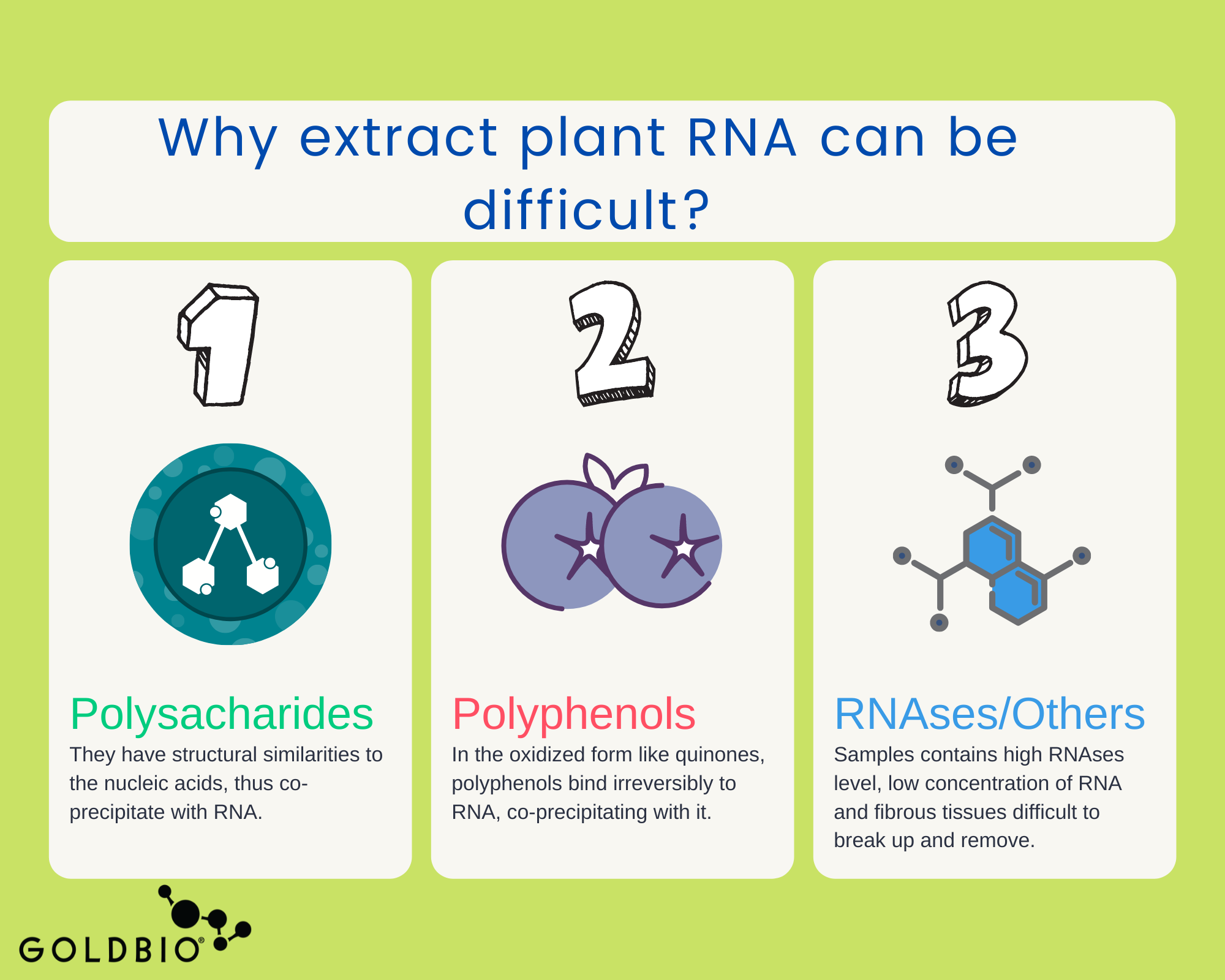
The trouble with plant RNA is that extraction generally produces low RNA quality and yield.
As you may know, plants have a lot of metabolites in their cells. They have proteins, lipids, polysaccharides, and secondary metabolites, to name a few.
All of these metabolites become mixed during the RNA extraction due to the lysis step, where it is necessary to break up the plant membrane to release the RNA.
The difficulty in isolating good RNA is because of the abundant presence of those metabolites. They are structurally similar to nucleic acids and co-precipitate along with RNA, thus decreasing both the yield and quality of the extraction. In addition, secondary metabolites like oxidized polyphenols (as quinones) irreversibly bind to nucleic acids and act as inhibitors for downstream applications or may degrade the RNA.
Some other conditions that make plant RNA extraction more difficult include: high levels of RNases (which degrade RNA) that are inherent in many plant samples, low concentration of RNA (due a high-water content), and fibrous tissues such as lignin (wood) that are difficult to break up and remove.
Again, isolating high-quality RNA is essential for downstream applications.
RNA is quite useful in many different techniques to understand gene regulation and expression. RNA is used for traditional applications like:
For advanced approaches, RNA is used for:
Thousands of protocols have been published for plant RNA extraction. However, most of the reported protocols are based on three methods:
Most of the RNA extractions can be split into four steps:

Although there are many solutions that are used in RNA, in this article we select six reagents/solutions that are very important for most of the RNA extraction protocols.
Generally, research studies regarding RNA aim to analyze the mRNA because it encodes for proteins, and it is key for regulation and gene expression studies. However, when an RNA extraction is performed, all of the different types of RNA are extracted.
To quantify the RNA yield, the μg RNA/mg tissue can be calculated. However, the quality is evaluated based on the ribosomal RNA content, which is the main type of RNA present in the extraction. Consequently, mRNA quality is calculated indirectly through the rRNA estimation on a gel.
Let’s explain how this “indirect measure of RNA” is estimated.
Ribosomes in plants are composed of two units, the larger subunit named 28S rRNA and small subunit as 18S rRNA. When both of these two units are present on a gel, it is an indication of high-quality RNA. If the RNA is degraded, one of the bands may look smeared, or neither of the two bands may appear at all.
A NanoDrop spectrophotometer is an instrument used to verify the concentration and purity (or quality) of the RNA obtained. To assess the presence and purity of the extracted RNA, researchers use the ratio of absorbance at 230 nm, 260 nm, and 280 nm.
If the RNA is pure, the 260/280 ratio is expected to be around 2. However, if the ratio is appreciably lower, it is an indication of the presence of phenol, protein, or other contaminants which absorb strongly at or near 280 nm.
On the other side, the 260/230 ratio, a range between 2-2.2 is expected. If this value is considerably lower, it can indicate the presence of contaminants (e.g. carbohydrates, EDTA, guanidine isothiocyanate, and phenol all absorb at 230 nm). Ratios lower than expected could require additional cleaning.
The bioanalyzer is another instrument that allows a more accurate assessment of the RNA quantity and quality. In this system, an electrophoretic separation on microfabricated chips is used for RNA sample separation. Then RNA is detected via laser induced fluorescence detection.
The Bioanalyzer results are represented as a chromatogram where two prominent peaks corresponding to 28S and 18S ribosomal units are an indication of pure RNA. Furthermore, the RNA Integrity number (RIN) is a quantitative measure of the RNA quality. An RIN between 7-9 is expected for a high-quality RNA. A guide to interpreting bioanalyzer results can be found here.
On the web, there are a lot of scientific forums with many researchers reporting their RNA extractions issues, and there are many requests for tips for getting successful RNA extractions. Below is a summary of tips to help in any RNA extraction protocol.
Make sure that every step of the RNA extraction process, and all of the tools used in the process, is kept cold. This is absolutely necessary in order to inhibit any RNase activity.
Be careful when using non-filter pipette tips because the inner part of the pipette barrel can be a source of contamination. Filter tips block particulate matter from contaminating your samples.
Bare hands are the main source of RNases. Wear gloves at all times, and make sure to change them frequently.
Although RNase/DNase free water is sometimes available for purchase, you can also prepare a solution with DEPC, which functions to inactivate RNase enzymes in order to avoid RNA degradation. There are RNases everywhere, so be careful to prepare all solutions with DEPC-treated water.
Although this tip may appear logical, it is important to dedicate enough time to read and search more information about your RNA protocol and specific plant species. This extra research can help you to detect bottlenecks and avoid making mistakes during the extraction process. Some plant species can be more challenging than others due to their chemical composition, such as polysaccharide or polyphenol concentrations, among others.
Often, you will start by following a written protocol and check things off as you go. However, during the process you may also make changes. Recording those changes is vital to understanding your results and helps you to optimize your protocol further. This will also help serve as a tool for others in your lab when they eventually need to do an RNA extraction.
Sometimes, depending how challenging your plant species is, additional cleaning may be required. For instance, if your sample has a lot of polysaccharides, a chloroform wash could help in the removal. Or if your sample has a lot of protein contaminants, an additional isopropyl alcohol wash could be used.

Ni2+ ions give nickel agarose beads their characteristic blue color. This blue color can fade or disappear completely when loading his-tagged proteins onto the column....

Nickel agarose beads change from blue to a brown or black color when the nickel ions have been reduced from a Ni2+ to a Ni1+...

The GoldBio Floating Tube Rack is one of our more clever giveaways because of the unique purpose it serves. And, with it also being one...

The characteristic blue color of nickel agarose beads comes from the 2+ oxidation state of the nickel ions. Color is also a useful indicator for...
