How Much Streptavidin Resin Should I use? Binding Capacity has the Answer
by Simon Currie, Ph.D.

by Simon Currie, Ph.D.
en using streptavidin agarose beads to purify biotinylated nucleic acids or proteins, you’ll need to decide exactly how much resin to use for your purification. To make this decision, you’ll estimate or calculate how many biotinylated molecules you’re purifying and use the binding capacity of the agarose beads to determine how much streptavidin resin to use.
The binding capacity of GoldBio’s streptavidin agarose beads is greater than 120 nanomoles of free biotin per milliliter (mL) of resin. The exact binding capacity will depend on the specific biotinylated molecule you are purifying and your buffer conditions.
We discuss in more detail what exactly agarose bead binding capacity is, and how to use it to estimate the amount of resin you need in this article about the binding capacity of Protein A, Protein G, and Protein L agarose beads. Three pertinent points to reiterate here are:
In this article we’ll discuss these factors, how to estimate how much agarose bead resin you need, and how to adjust the amount you use for the next purification, if necessary, based on your results.
How does biotinylated molecule size impact binding capacity?
How does buffer influence binding capacity?
Biotinylated molecules such as nucleic acids, proteins, and antibodies are frequently purified using streptavidin agarose beads. An important step in the purification process is binding the biotinylated molecules to the streptavidin beads during the loading step (Figure 1).
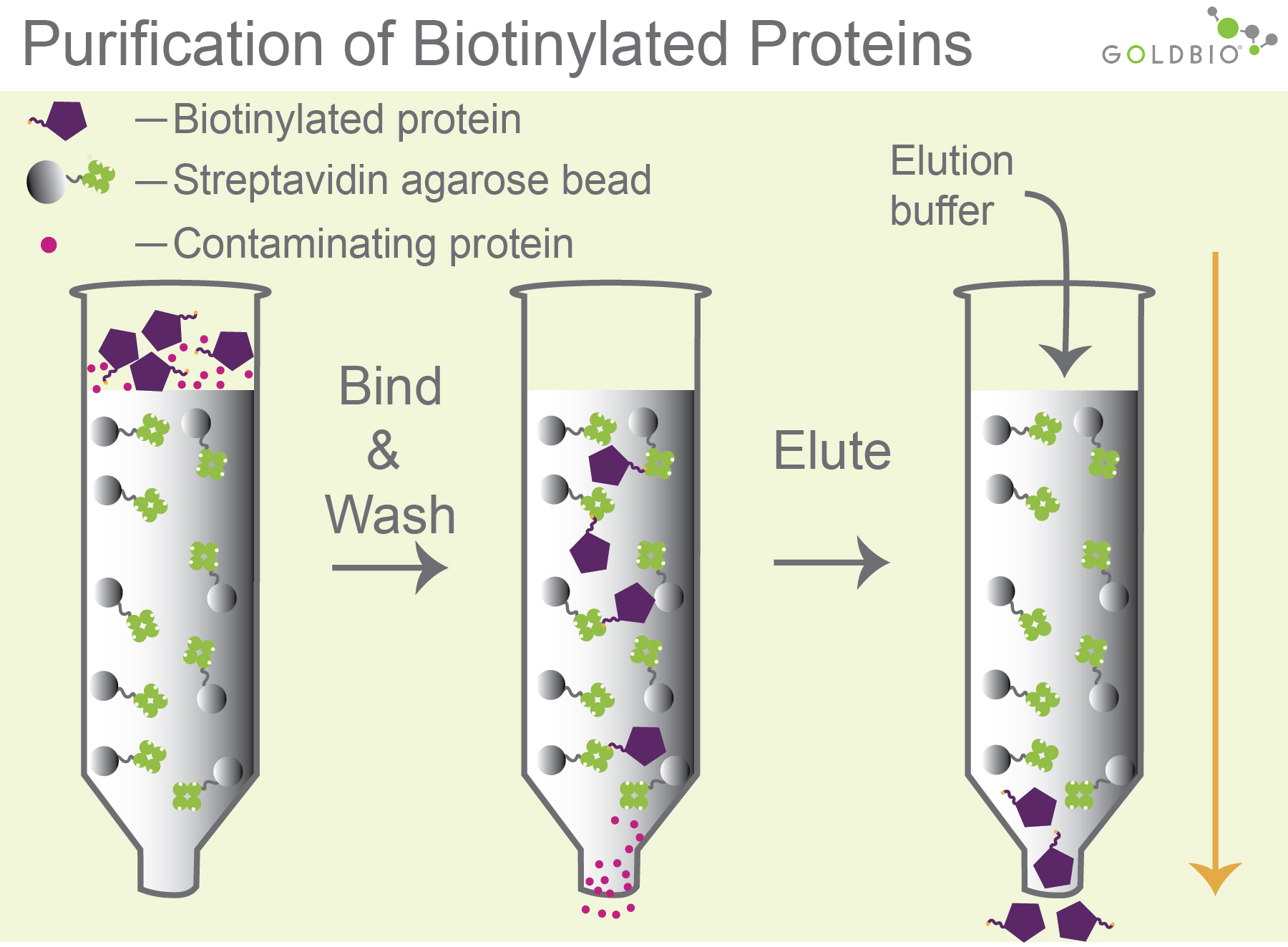 Figure 1.
Purification of biotinylated molecules. Biotinylated molecules bind to streptavidin
agarose beads (column 2). After washing, the biotinylated molecules
can be eluted a number of ways, including with an acidic pH elution buffer that
weakens the interaction between the streptavidin and biotin (column 3).
Figure 1.
Purification of biotinylated molecules. Biotinylated molecules bind to streptavidin
agarose beads (column 2). After washing, the biotinylated molecules
can be eluted a number of ways, including with an acidic pH elution buffer that
weakens the interaction between the streptavidin and biotin (column 3).
So, how do you know how much streptavidin agarose beads to use for your purification? If you’re doing batch chromatography using gravity columns, this generally means roughly a few milliliters of the agarose bead resin. Knowing the binding capacity of streptavidin agarose beads will help you better estimate how much resin to use for your purification.
The binding capacity of a particular type of agarose bead is how much of a given molecule that bead can bind to. GoldBio’s streptavidin agarose beads bind to greater than 120 nanomoles of free biotin per mL of resin.
Since the binding capacity refers to mL of resin, it is important to know what this term means. Like most agarose beads, streptavidin beads are shipped as a 50% slurry, meaning the solution is 50% agarose beads and 50% liquid solution. So, you’ll want to invert the bottle several times to make sure that the slurry is as homogenous as possible and there are not any big clumps of agarose beads stuck at the bottom of the bottle.
Then, you’ll pour twice the volume of desired resin into your column. So, for example if you want 1 mL of agarose bead resin for your purification, you’ll pour 2 mL of mixed slurry into your column, then let the 1 mL of buffer run through leaving you with 1 mL of agarose bead resin (Figure 2).

Figure 2. The volume of the settled agarose resin (right) will be about half of the 50% slurry that you put into the column (left).
You may have noticed that the above stated binding capacity for streptavidin beads refers to free biotin. In all likelihood you won’t be purifying free biotin, but rather a biotinylated molecule such as a protein or nucleic acid. The size of your biotinylated molecule will influence the binding capacity on agarose beads.
The bigger the biotinylated molecule is, the lower the binding capacity of streptavidin agarose beads will be. Let’s walk through why exactly that is the case.
Streptavidin is a tetramer, meaning that four copies of the streptavidin bind to each other, and are able to bind up to four biotin molecules per streptavidin tetramer. Free biotin is small, roughly one thousand times smaller than streptavidin, so free biotin can bind to all four binding sites on streptavidin without the biotin molecules running into each other. Similarly, for small biotinylated molecules, they can also interact with every streptavidin binding site (Figure 3).
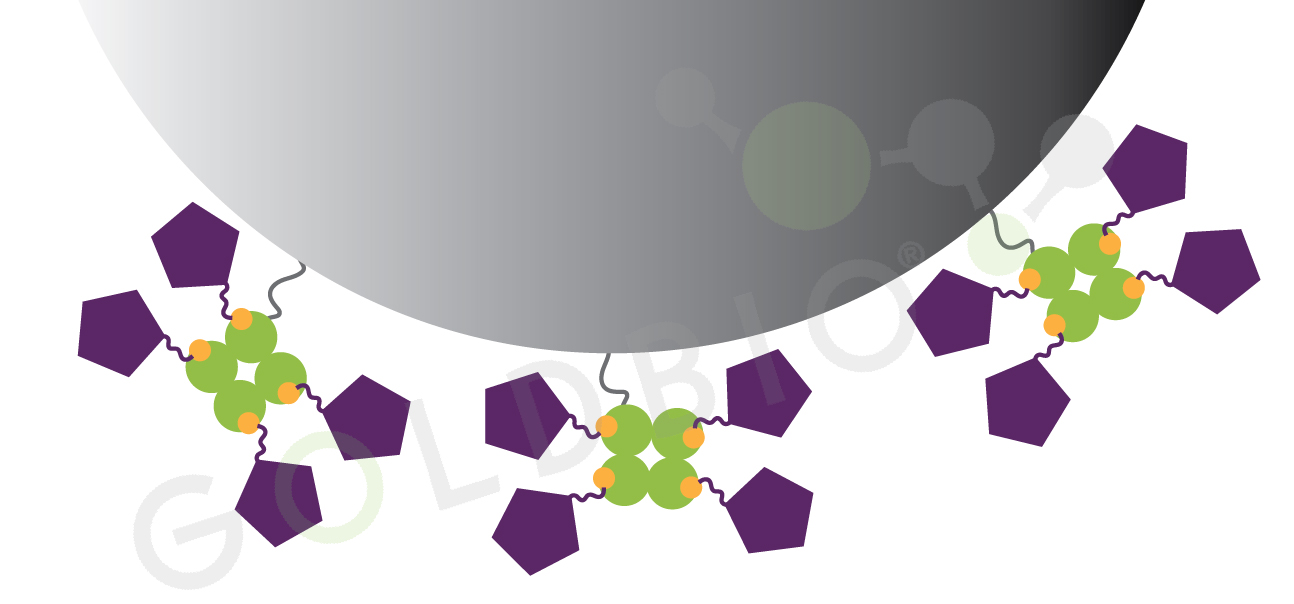
Figure 3. Binding capacity is higher for smaller biotinylated molecules. Smaller biotinylated molecules can bind to every streptavidin site without being blocked by each other or by the agarose bead.
But now, let’s assume you are purifying a really big
biotinylated protein. The biotinylated protein may not be able to bind to all
four binding sites on streptavidin because once one of the big proteins is
bound, it will physically block other big proteins from binding to neighboring
sites on streptavidin (Figure 4).

Figure 4. Binding capacity is lower for bigger biotinylated molecules. Larger biotinylated proteins block each other from binding to streptavidin beads, so not every streptavidin binding site is occupied.
So, keep this in mind when estimating how much streptavidin agarose beads to use. If you’re purifying a pretty small biotinylated molecule, then you can probably get pretty close to the 120 nanomoles per mL binding capacity. If you’re purifying a big biotinylated molecule, then the binding capacity will be a fraction of that for your molecule of interest.
In practice, there is not a binary threshold that separates molecules into small and big. But keep in mind the sizes of biotin (~ 0.2 kDa) and streptavidin (~ 54 kDa). Small biotinylated molecules, let’s say around 10 kDa and smaller, should bind to streptavidin with high efficiency. As your biotinylated molecule approaches, and exceeds, the size of streptavidin you would expect to have lower binding capacity due to steric hindrance between your biotinylated molecules and the agarose beads.
The binding capacity of streptavidin agarose beads will depend on the buffer you are using to load antibodies onto the column. To consider an extreme example,if you use an elution buffer as your loading buffer, the binding capacity of these beads will be close to zero – so definitely don’t do that! Typically, using a loading buffer with neutral pH and lower salt, like PBS for example, will help biotinylated molecules bind to the streptavidin beads with near optimal binding capacity.
Purification is an iterative process, and you usually won’t get it perfectly right the first time. If you don’t capture all of your biotinylated molecule, you used too few beads and should scale up next time. If you captured all of your biotinylated molecule you may be able to reduce the volume of streptavidin beads that you use next time to help your research dollars stretch further. See this article for more details about evaluating how much agarose bead resin to use.

IPTG and auto-induction are two ways to induce protein expression in bacteria. They work similarly, but have different trade-offs in terms of convenience. While IPTG...

The final concentration of IPTG used for induction varies from 0.1 to 1.0 mM, with 0.5 or 1.0 mM most frequently used. For proteins with...

A His-tag is a stretch of 6-10 histidine amino acids in a row that is used for affinity purification, protein detection, and biochemical assays. His-tags...

Competent cells such as DH5a, DH10B, and BL21 will maintain their transformation efficiency for at least a year with proper storage. It is important to...
