What is a GST Pull-Down Assay? The Complete Overview with Protocols and Analyses Guides
by Simon Currie, Ph.D.

by Simon Currie, Ph.D.
Molecular interactions are foundational for homeostasis at cellular and organism levels, and the dysregulation of these interactions causes many diseases.
That’s why when a scientist discovers a particular protein that’s important in a cellular process or disease, one of the first things they will do is examine its molecular interactions. There are a few ways to investigate protein-protein interactions. One powerful and commonly used approach is to tag your protein of interest with Glutathione S-Transferase (GST) and perform a GST pull-down to see what binds to the protein of interest.
GST is a solubility tag that is also used to identify and verify protein-protein interactions. By tagging their protein of interest with GST, researchers can then immobilize it on glutathione agarose beads, and flow over a complex, or simple biochemical mixture to identify interacting proteins.
GST pull-down assays are used to identify protein-protein interactions. A GST-tagged protein is immobilized on glutathione agarose beads and used to pull down interacting proteins from a complex mixture, such as cell lysate, or using purified recombinant proteins.
In this article we’ll discuss how GST-fusion proteins and glutathione agarose beads are used to perform GST pull-down assays in order to identify protein-protein interactions.
What is a GST pull-down assay?
GST pull-down vs immunoprecipitation
Preparing your glutathione beads
Loading your GST-fusion protein onto the glutathione beads
Adding the interacting proteins
Eluting the GST-fusion and interacting proteins
Materials needed for GST pull-down assays
Preparing glutathione agarose beads
Loading GST-fusion proteins onto glutathione beads
NOTE: By completing this form, you're opting in for our emails about helpful resources, GoldBio sales, and new products. And you can always opt out anytime by clicking "unsubscribe" at the bottom of a promotional message. Please review GoldBio's privacy policy.
GST pull-downs are assays used to investigate protein-protein interactions. For this approach to be useful, you need to know the identity of one of the proteins that you’re interested in investigating. Then, you’ll clone an expression construct that fuses GST to your protein of interest and purify your GST-fusion protein.
Now your GST-fusion protein is ready to be immobilized on glutathione agarose beads to see what interacts with it.
You don’t have to know what your protein of interest interacts with beforehand. In fact, many GST pull-downs use cell lysates to try to discover new interactions with their protein of interest (Figure 1). For example, let’s suppose that your protein of interest is important in lung cancer. You could use lysates from lung cancer cells to try to identify novel interactions in this relevant cell type.
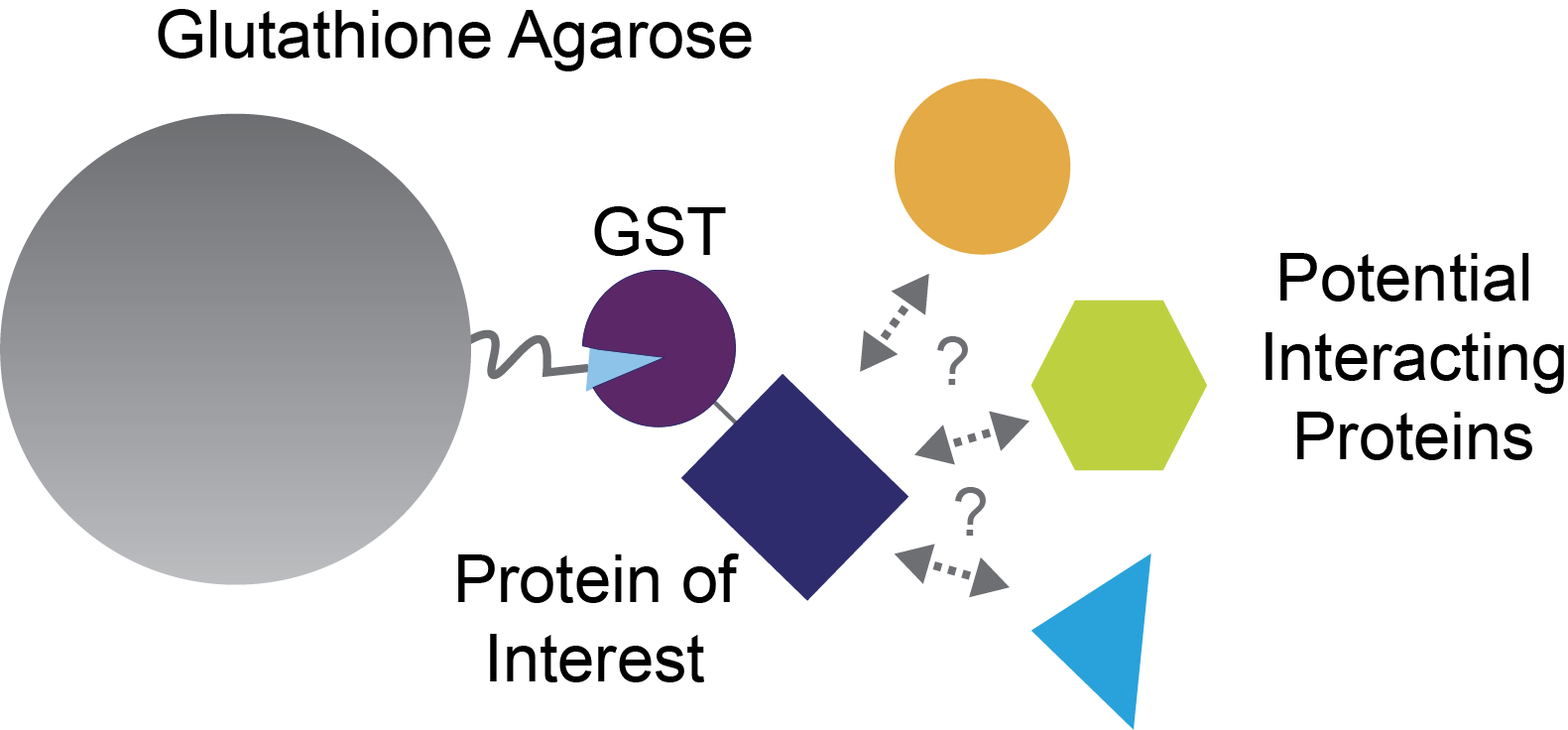
Figure 1. GST pull-down assay for investigating protein-protein interactions.
Even if you do already know an interaction with your protein of interest, GST pull-downs can also be useful in learning more detailed information about this interaction, as we’ll discuss below.
GST pull-downs are conceptually very similar to immunoprecipitation assays in that both techniques are used to identify protein-protein interactions. The only difference is how your protein of interest is connected to the agarose beads, which has important implications for how you elute it, and any proteins that it binds to from the column.
In immunoprecipitation, Protein A, Protein G, or Protein L agarose beads are used to bind to an antibody that recognizes your protein of interest. For GST pull-downs, however, the GST in your GST-fusion protein binds to glutathione agarose beads (Figure 2).
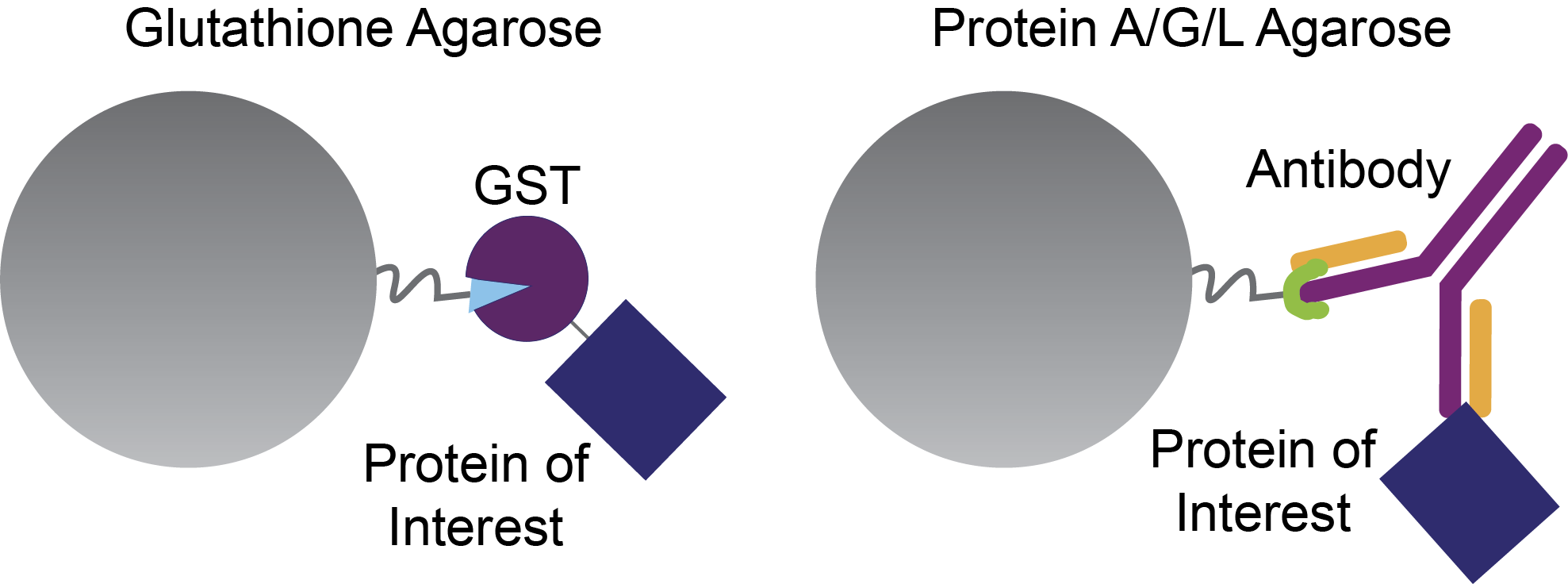
Figure 2. Glutathione agarose beads bind GST-fusion proteins (left), and Protein A, G, and L beads bind to antibodies. Both of these setups are used to investigate the interactions of a protein of interest – the blue rectangle.
As we’ll discuss in the elution section below, eluting GST-fusion proteins from glutathione agarose beads is gentle for most proteins compared to the harsh elution conditions required for Protein A, G, and L beads.
Additionally, pull-downs with glutathione beads tend to have less nonspecific binding relative to using nickel agarose beads with his-tags.
For these reasons, it can be worth the extra effort to clone an expression construct for a GST-fusion form of your protein of interest if you’re searching for interacting proteins.
Table 1. Comparing GST pull-downs and immunoprecipitation assays.
|
Feature |
GST Pull-Down |
Immunoprecipitation |
|
Binding Moiety |
GST Tag |
Antibody (Protein A/G/L) |
|
Beads |
Glutathione Agarose Beads |
Protein A/G/L Agarose |
|
Elution Harshness |
Mild |
Harsh |
|
Specificity |
High |
Varies |
|
Experimental Requirement |
Cloning with GST Tag |
Antibody availability |
There are 6 main steps to conduct a GST pull-down:
We’ll cover each of these steps in more detail in the subsections below.
GoldBio’s glutathione agarose beads come as a 75% v:v mixture in 20% ethanol. You won’t want to expose your protein of interest or the interacting proteins to 20% ethanol, so after adding your beads to a plastic column you’ll first rinse the beads out with deionized water (dIH2O). Then you’ll equilibrate your beads in the binding buffer, or the same buffer that you’ll load your GST-fusion protein onto the glutathione beads, as we’ll discuss in the next section.
One key consideration for preparing your glutathione agarose beads is how much volume of beads you need to use for your pull-down. GoldBio’s glutathione agarose beads have a binding capacity of up to 8 milligrams (mg) of GST-fusion protein per milliliter (mL) of beads. So, if you know how much of your GST-fusion protein you’re going to load onto the beads, it will be very easy to calculate the volume of beads that you need.
Glutathione bead preparation steps:
An important consideration for loading your protein onto the glutathione beads is what binding buffer to use. In our official GST affinity purification protocol we recommend using 1x PBS, pH 7.3 as the binding buffer. This buffer has a neutral pH, physiological salt concentrations, and works great for a lot of proteins.
However, glutathione agarose beads are pretty compatible with a wide range of buffer pHs and additives. So if you know your protein of interest is happy in another buffer, you can likely use that for the binding buffer. Just check this list first to make sure that you don’t have any buffer components that would be incompatible with GST capture by the glutathione beads. And definitely don’t add any glutathione to your binding buffer – save that for the elution step.
To load your protein onto the glutathione agarose beads, first cap the bottom of your plastic column, then add your protein of interest, and cap the top the column. Use gentle agitation such as rotating or shaking at a low speed to mix the beads and your protein of interest. As much as possible, try to match the buffer that your interacting protein source is in with your binding buffer. Remember, there’s quite a bit of flexibility here so if you need to use a buffer besides PBS to accommodate your interacting protein, just use that one for the binding buffer as well.
Relative to other affinity interactions, the binding between GST and glutathione is pretty slow, so you’ll want to incubate your protein of interest with the beads for around 30 minutes to an hour before uncapping the column and letting the flow-through drip out the bottom. You can collect this flow-through with tubes in case you want to analyze the binding efficiency. Most of your GST-fusion protein should be on the beads, but if something went wrong with your buffer you would have collected the GST-fusion protein and could try again.
It’s good practice to wash your column with a couple column volumes of binding buffer to remove any unbound proteins.
Loading GST-fusion proteins onto glutathione beads steps:
Now that you have your protein of interest bound to the glutathione agarose beads through GST, you’ll want to add the potential interacting proteins to this mixture.
Typically, this step will happen in one of two ways. If you’re trying to identify novel interactors with your protein of interest, then you’ll probably use a complex biochemical mixture like cell lysate, for example.
Alternatively, if you already have a specific protein in mind and you want to see if it interacts with your protein of interest, then you would have purified recombinant proteins ready to add to the glutathione beads.
Whatever your protein source is, you’ll want to load it onto the column in pretty much the same way – cap the bottom, add your interacting protein or cell lysate, cap the top, and incubate with gentle agitation for around an hour.
After incubation, uncap the column and let the flow-through fraction flow out the bottom. Collect this sample and save it for the analysis step.
Then wash the column with at least 2 column volumes of binding buffer to remove any unbound proteins. Also collect this sample and save it for the analysis step (Figure 3).
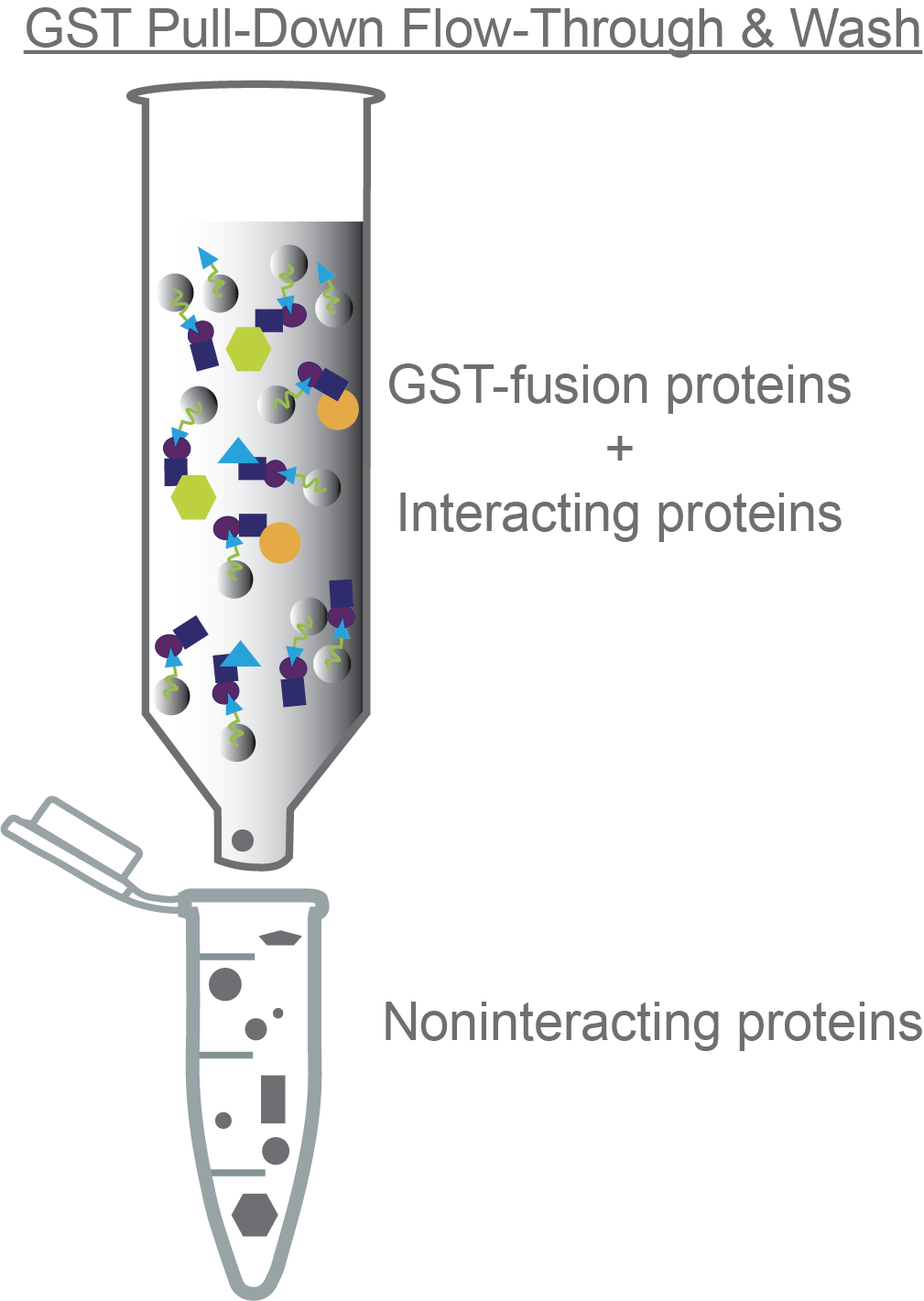
Figure 3. Non-interacting proteins will come through the column during the flow-through and wash steps.
Adding interacting proteins onto glutathione beads steps:
Now you’re ready to elute your GST-fusion protein and any interacting proteins from the glutathione column. To do this, you’ll add your elution buffer. Like the binding buffer, the elution buffer is pretty flexible in terms of buffer pH, salt, additives, etc. Importantly, your elution buffer needs to contain ~ 10-20mM reduced L-glutathione. The free glutathione will bind to your GST-tagged protein of interest, freeing it and any interacting proteins from the agarose beads.
Add the elution buffer to the column, cap both ends, and incubate for a few minutes before uncapping and dripping your elution out of the bottom (Figure 4). Collect these fractions – this is where you’re expecting your protein of interest and any interacting partner proteins to be.
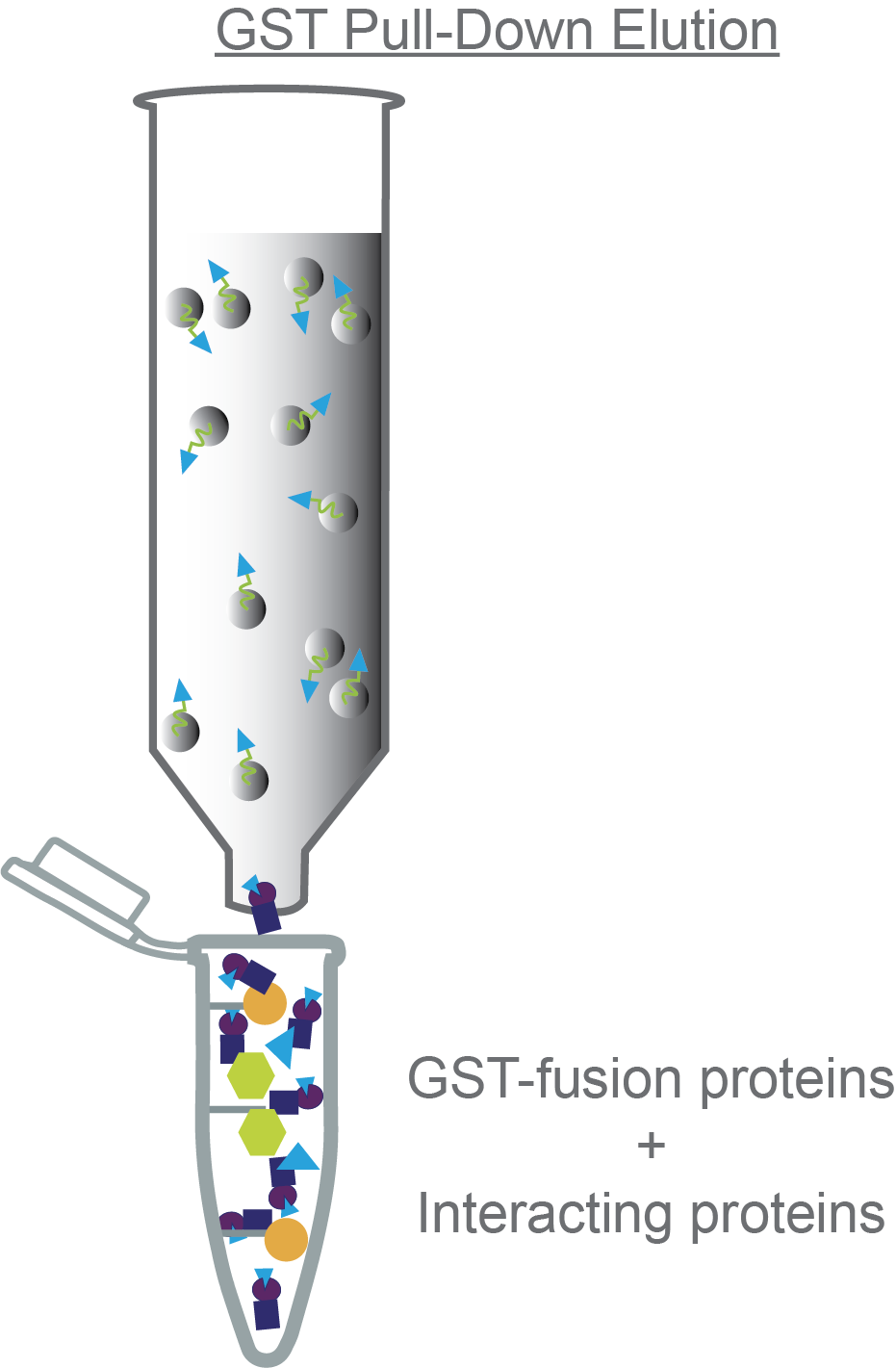
Figure 4. Eluting the GST-fusion protein and any interacting proteins from the column.
Eluting GST-fusion proteins and interacting proteins steps:
This section is a quick summary of the material you’ll need for each core step of a GST pull-down assay. For each subsequent step, only additional supplies are listed while previous supplies like plastic columns and beads are assumed to carry over from the previous step.
Now that you’ve done the actual GST pull-down experiment, it’s time to analyze the results.
Usually, the first way to analyze the pull-down is to run an SDS-PAGE gel on the samples you collected.
An SDS-PAGE gel can help you answer questions such as:
First let’s consider a scenario where you’re using cell lysate in your pull-down experiment to identify novel interacting proteins. Are there any protein bands in your elution other than your GST-fusion protein? If yes, then those proteins interact with your protein of interest. The hypothetical SDS-PAGE gel in Figure 5 shows 3 interacting proteins.
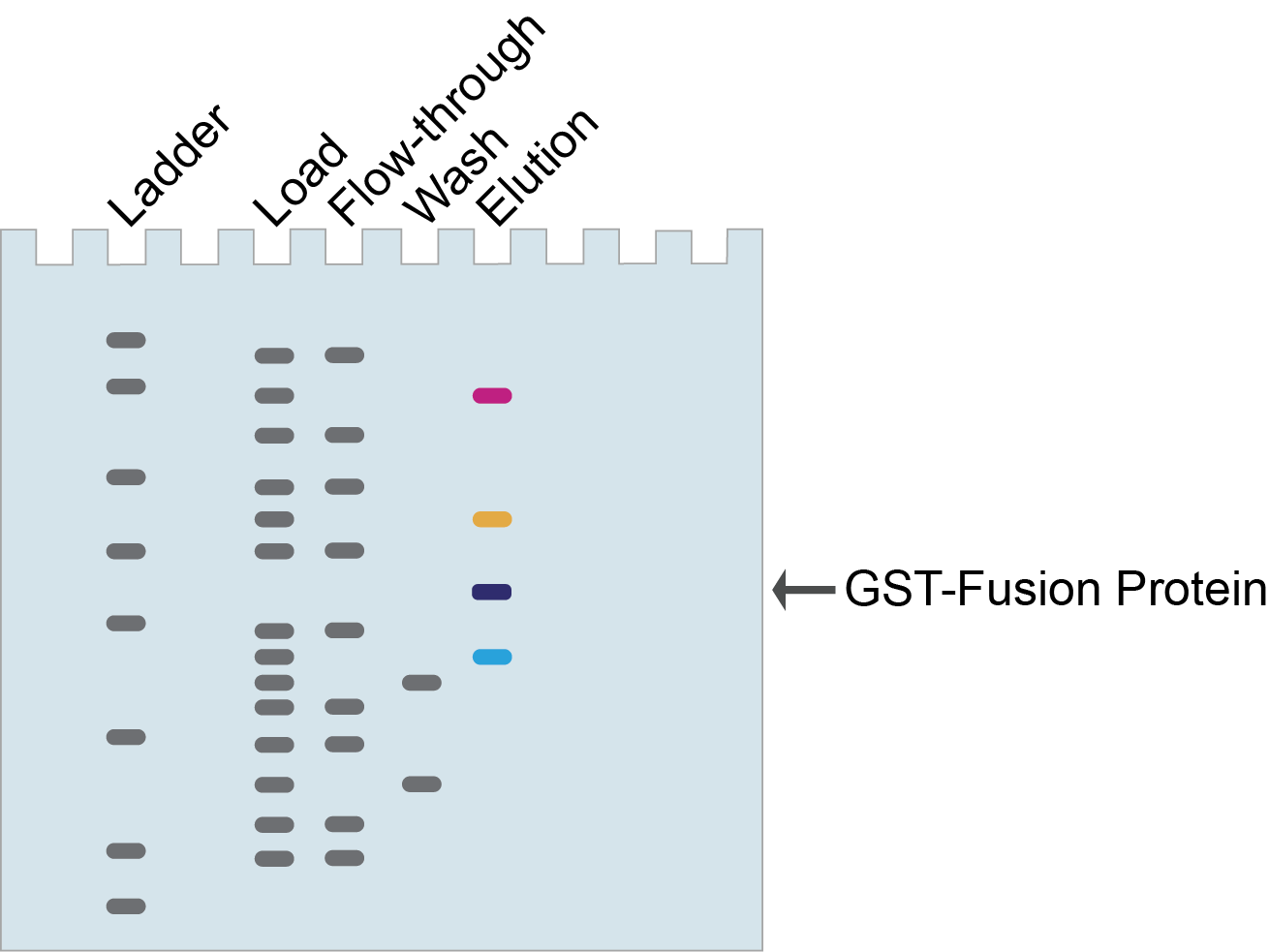
Figure 5. Identifying novel interacting proteins with GST pull-downs.
In Figure 5, the proteins (pink, orange, and blue bands) that co-elute with your GST-fusion protein (purple band) are candidate interacting proteins and can be further identified and validated.
Usually, the next step is to use mass spectrometry to identify those interacting proteins (Kassa et al, 2023). This is a great approach because it is unbiased and doesn’t require any previous knowledge or guessing.
However, if you think you know which proteins might be interacting with your protein of interest, perhaps based on their size and biological context, then you could do a Western blot using antibodies against the proteins you think they are.
Ok, now let’s consider a scenario in which you already have a candidate interacting protein, and want to determine which part of it interacts with your protein of interest (Figure 6). So, you would have already purified proteins corresponding to the full-length and each individual domain of the interacting protein, and now you do a pull-down to see which ones bind.
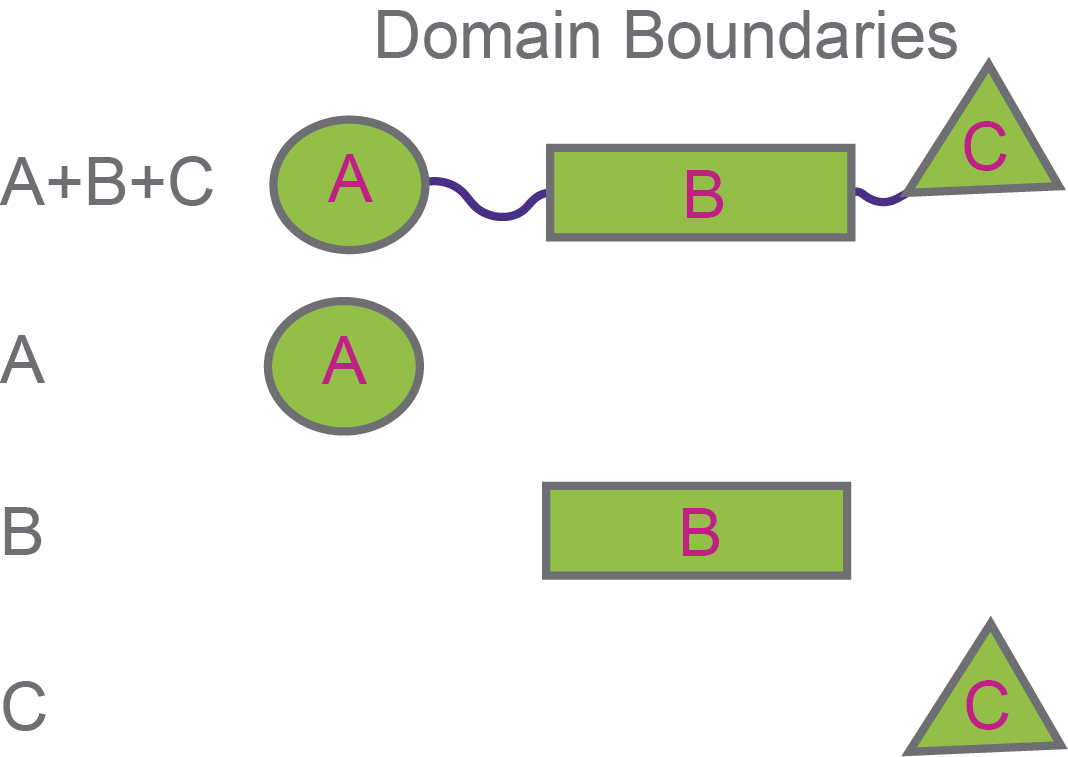
Figure 6. Testing different domains of an interacting protein to see which one binds to your protein of interest.
Figure 7 shows some hypothetical SDS-PAGE gels for GST pull-downs with these different domains. In this example, the oval and rectangle domains are only in the flow-through, but not in the elution so they do not interact with your protein of interest.
In contrast, the triangle domain (and the full-length protein) is “pulled-down” by your protein of interest so they co-elute.
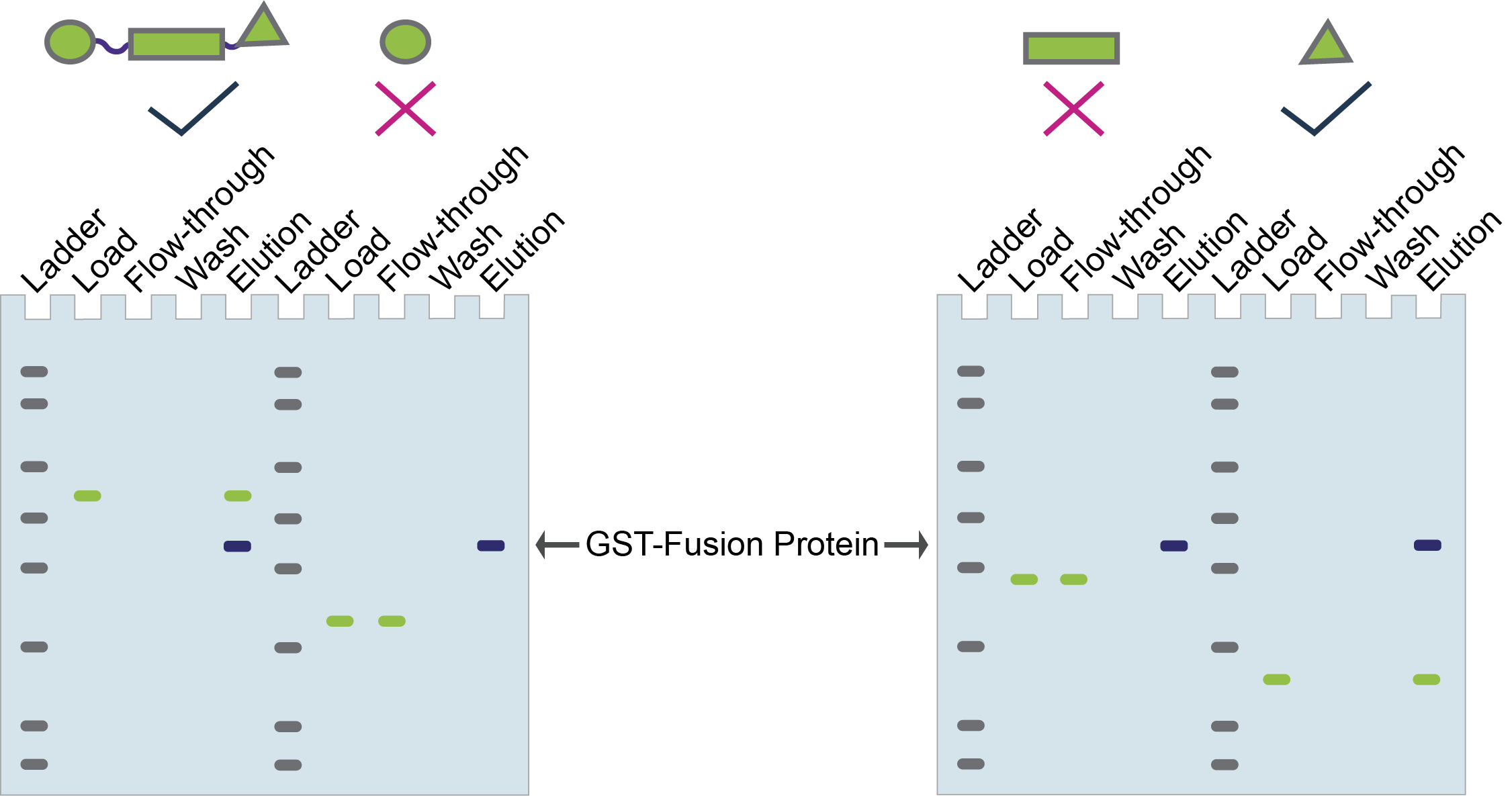
Figure 7. Hypothetical GST pull-down to identify the domain of a protein that interacts with your protein of interest.
Oftentimes, when doing pull-down experiments, researchers will include a control experiment using GST alone instead of their GST-fusion protein. If the interacting protein that you’re analyzing also binds to the GST only control, then this may be through non-specific interactions to the beads or to GST (Figure 8).
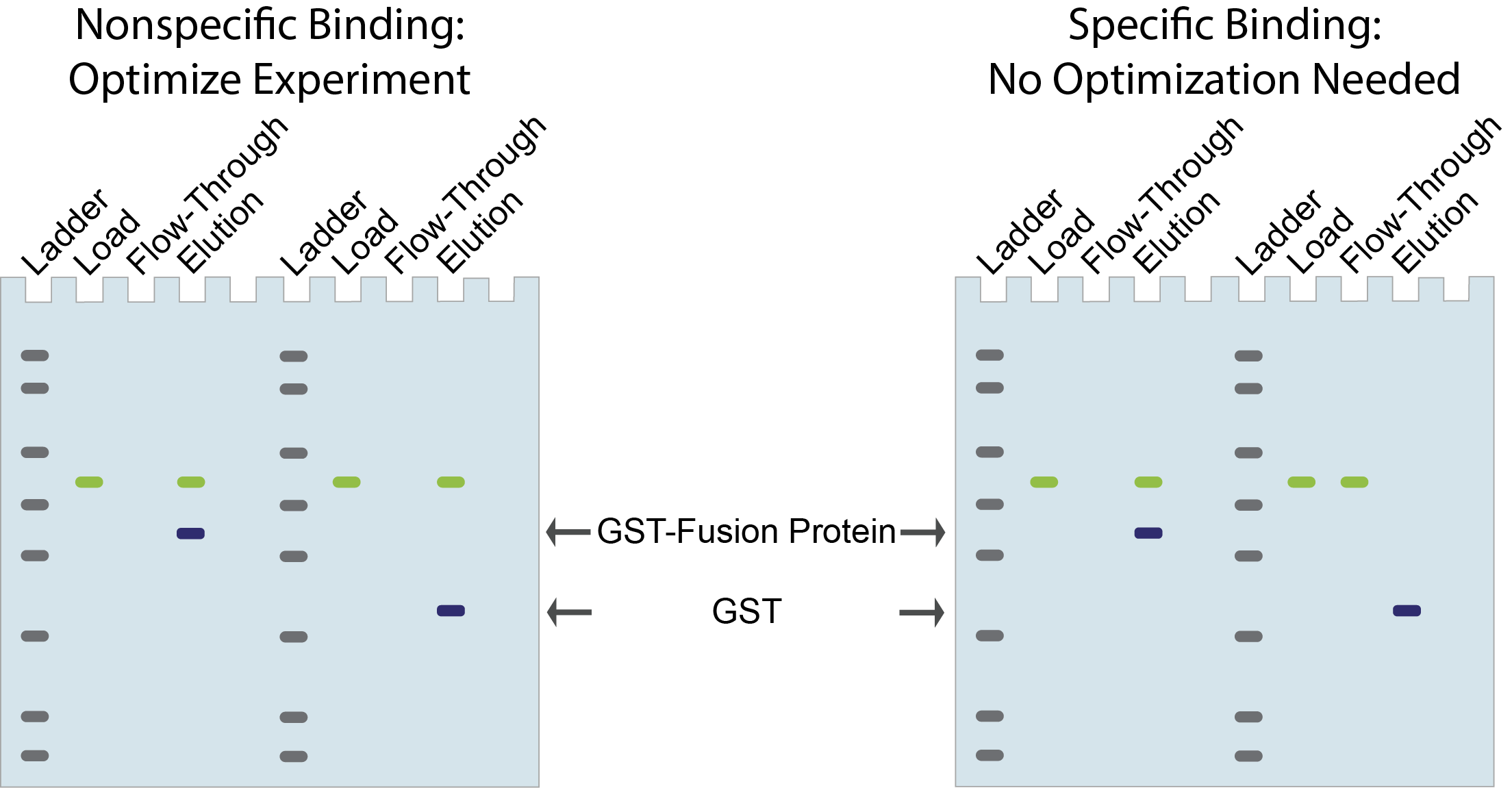
Figure 8. GST
only control for pull-downs to determine specificity of interacting proteins.
Sometimes this issue is fixed simply by optimizing buffers, for instance by increasing salt or detergent concentration, so that the interacting protein no longer binds to the GST only control, but still binds to your GST-fusion protein of interest.
If you’re not able to quickly optimize the buffer conditions to see this separation, then the interacting protein may not be a “bona fide” interactor and something that you want to spend more time investigating.
So that’s how you can use GST pull-downs to investigate interactions with your protein of interest.
At GoldBio we have lots of great resources for learning more about glutathione agarose beads, and plenty of research reagents to aid in your pull-down assays. Whether you’re still learning about this technique or ready to do GST pull-downs in your lab, check out the helpful links to GoldBio products and resources below.

Ni2+ ions give nickel agarose beads their characteristic blue color. This blue color can fade or disappear completely when loading his-tagged proteins onto the column....

Nickel agarose beads change from blue to a brown or black color when the nickel ions have been reduced from a Ni2+ to a Ni1+...

The GoldBio Floating Tube Rack is one of our more clever giveaways because of the unique purpose it serves. And, with it also being one...

The characteristic blue color of nickel agarose beads comes from the 2+ oxidation state of the nickel ions. Color is also a useful indicator for...
